23 September 2017
With an organ as complex as the brain, there are numerous ways in which even a minor mutation can cause profound developmental problems. A genetic analysis of children born with neurological problems now reveals how various genetic disruptions effectively disable a protein with a prominent role in brain formation.
A protein called DYRK1A helps regulate the expression of key genes involved in nervous system development. The loss of even one of the two copies of the gene encoding it can have severe neurological consequences, including seizures, autism and intellectual disability. To better understand the importance of this protein, researchers led by Caroline Wright of the Wellcome Trust Sanger Institute in the UK combed through genetic data collected by the Deciphering Developmental Disorders (DDD) study.
The DDD study, coordinated by Wright, profiled thousands of children with neurodevelopmental disorders and their unaffected parents in order to home in on potential mutations causing them. The researchers found that 0.5% of these children carried mutations in the gene encoding DYRK1A. All of these children had intellectual disabilities, and the vast majority also had notable physical developmental problems including microcephaly — an abnormal shrinkage of the skull.
Wright’s team then profiled the various DYRK1A mutations these children carried. Fourteen out of the 19 were so disruptive as to effectively terminate production of the protein altogether. But the other five were more subtle mutations and the researchers were subsequently able to map these to the various functional domains of DYRK1A.
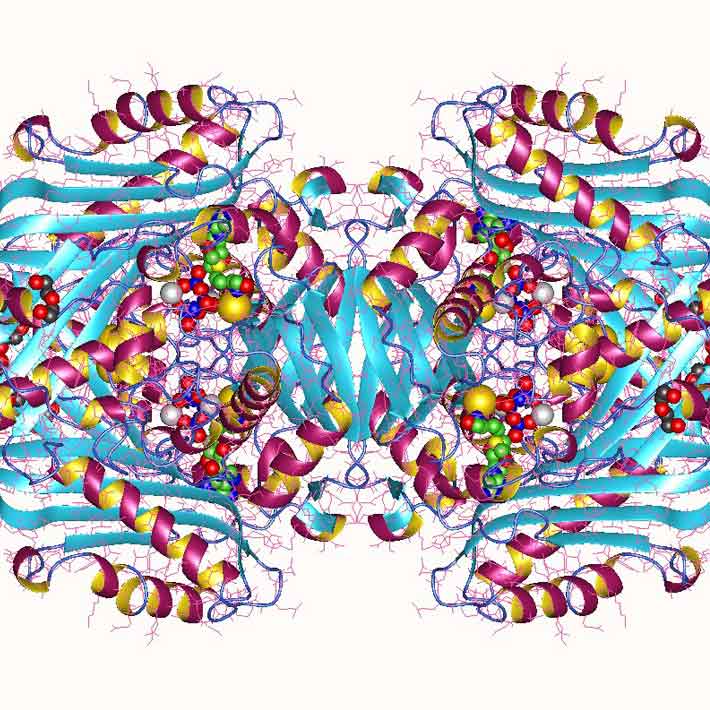
DYRK1A is an enzyme that transfers a phosphate chemical group from a molecule called ATP onto other target proteins. All five mutations in some way affected the ‘kinase domain’ of the enzyme that facilitates this transfer. Some mutations had obvious effects, such as disrupting ATP binding, and these were associated with more severe disability. In contrast, other mutations that exerted less direct effects on DYRK1A function by slightly destabilizing the protein’s structure were also associated with milder impairments.
The researchers subsequently examined a large number of other previously reported variants in the DYRK1A gene. Notably, gene variants with no ill effects were far less likely to occur within the kinase domain than those associated with developmental problems. The details of DYRK1A function are still poorly understood, but these results reveal a critical role for this enzyme’s kinase activity in brain development, and the authors propose that similar mutations in related kinases might contribute to other diseases as well.
References
Evers, J.M., Laskowski, R.A., Bertolli, M., Clayton-Smith, J., Deshpande, C. et al. Structural analysis of pathogenic mutations in the DYRK1A gene in patients with developmental disorders. Hum. Mol. Genet (2017).| article