20 December 2021
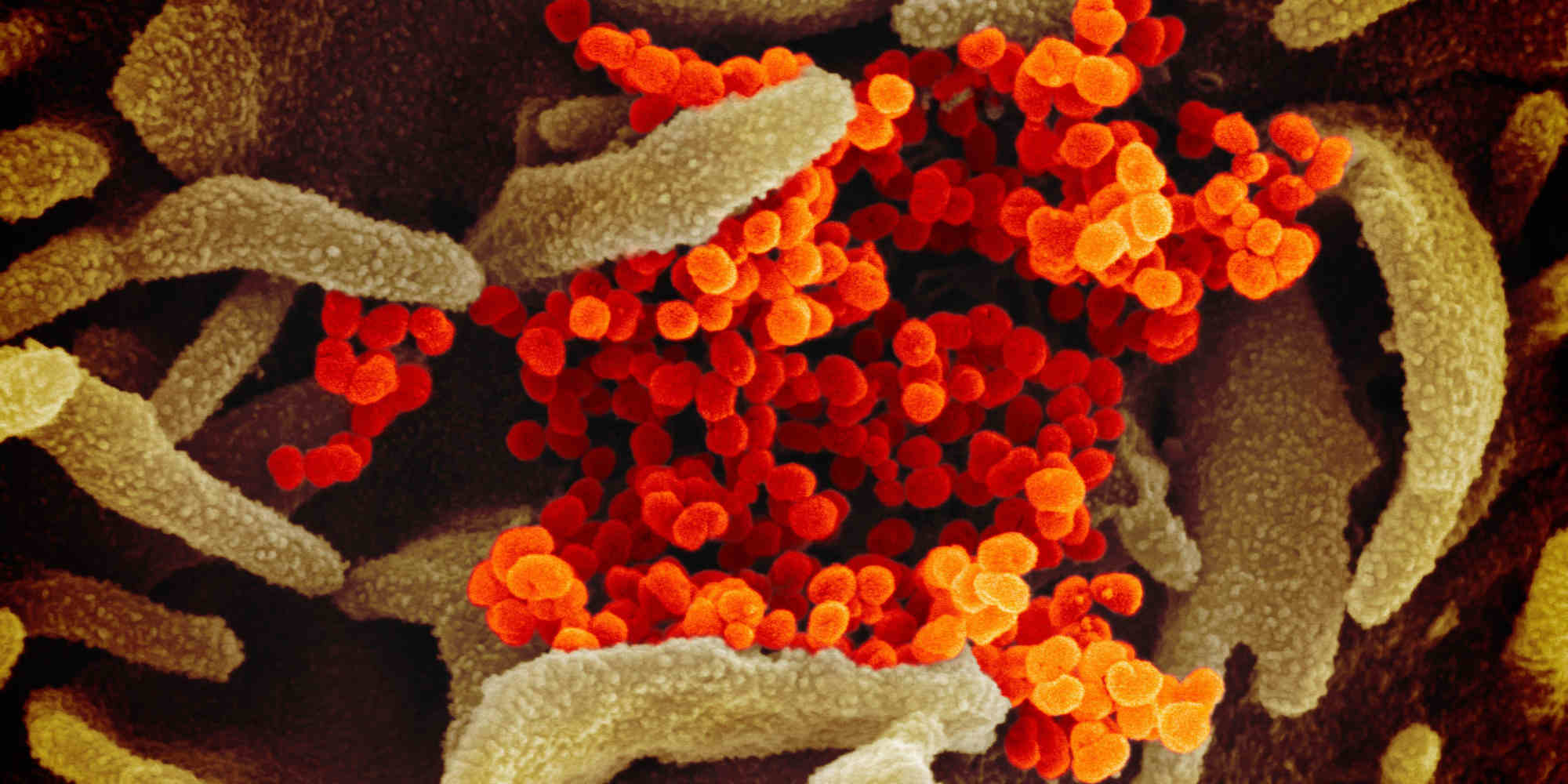
Molecular level insight into the evolutionary pathways and structure of SARS-CoV-2, the virus responsible for COVID-19, could help manage present and future coronavirus-related epidemics, suggest researchers at King Saud University.
SARS-CoV-2 has spread exponentially, causing a global pandemic that continues to affect millions of people worldwide. The novel coronavirus is genetically similar to other human viruses that cause severe acute respiratory syndromes, such as SARS-CoV and MERS-CoV, as well as bat coronaviruses, suggesting that it originated from a bat-borne virus. The ongoing rise in infections has resulted in genetic modifications, producing multiple variants of the virus that have made the epidemic difficult to control.
Led by Saleh Eifan, the team has now devised a gene sequencing approach to better understand the emergence of these variants. The researchers identified the selection pressures that have driven changes in the gene sequence encoding the spike proteins. Spike proteins dot the SARS-CoV-2 surface and play a crucial role in infection by enabling the initial binding of virus particles and their fusion with host cells.
The researchers compared three SARS-CoV-2 spike protein sequences from Saudi Arabia to those from SARS-CoV that caused the recent SARS outbreak and from bat SARS-like coronaviruses collected between 2011 and 2017 in Yunnan province and elsewhere in China. They discovered seven mutations resulting from positive evolutionary pressure on the SARS-CoV-2 spike protein. Five of these alter amino acids in protein domains responsible for receptor binding. These mutations could enhance the receptor’s affinity and consequently the transmissibility of the virus, as observed in the SARS-CoV-2 variant B.1.1.7, known as Alpha.
Despite the strong overall resemblance between SARS-CoV-2 and bat SARS-like coronavirus, their spike proteins had several variations in their individual constituent chains. The researchers found that the SARS-CoV-2 spike protein has a more open configuration and multiple polysaccharides that could facilitate viral entry into host cells and provide a potential target for antibodies. Furthermore, the subunit responsible for transmembrane fusion contained a site that can be cleaved by proteases and can thus be investigated as a potential target for antiviral drugs.
The researchers are currently exploring ways to use these findings to develop therapeutic and preventative approaches against coronaviruses.
References
- Nour, I., et al. Molecular adaptive evolution of SARS-COV-2 spike protein in Saudi Arabia. Saudi Journal of Biological Sciences 28, 3325–3332 (2021). | article