10 June 2021
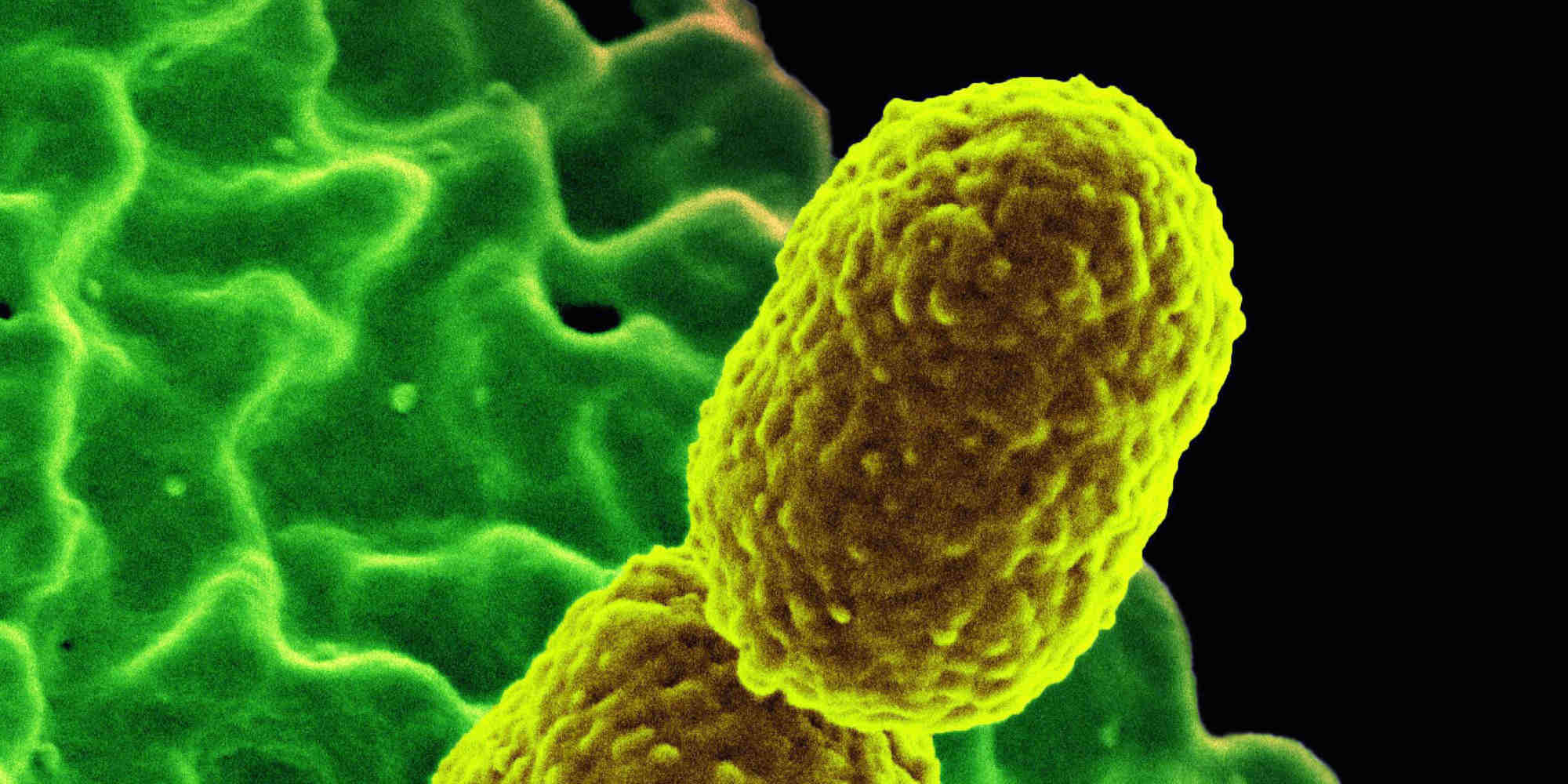
Bacterial pathogens such as Escherichia coli and Klebsiella pneumoniae are becoming increasingly resistant to antibiotics, with significant global health implications. Multiple antibiotics are available to treat K. pneumoniae infections, but the pathogen has evolved to evade almost all of them, even the carbapenem family of antibiotics that was considered a last line of defence.
Majed Alghoribi, chairman of KAIMRC’s Infectious Disease Research Department led a team that examined K. pneumoniae strains currently circulating in the National Guard Hospital in Riyadh to determine which specific resistance-conferring genes are present in the region. They also verified the genotypes of the predominant strains to facilitate control programs. Their findings will inform infection control intervention strategies and help promote stringent but effective antibiotic use.
Bacteria evolve rapidly via various mechanisms, such as mutations that alter aspects of their structure so that antibiotics can no longer attach to them and destroy them. Bacteria can also share DNA information with one another via fragments called plasmids, which can carry genetic codes that promote resistance.
Alghoribi and his team examined 50 K. pneumoniae isolates from different clinical sources taken from hospital patients. All were resistant to more than three antibiotics, including carbapenems. The researchers found that 49 out of the 50 isolates harboured the CTX-M gene, a common antibiotic resistant gene found in K. pneumoniae strains across the globe.
The team also found variants of the OXA gene, which is known to confer carbapenem resistance, in 43 of the isolates, and 16 samples harboured another carbapenem-resistance gene, NDM-1 They also identified two newer OXA phenotypes that were phylogenetically distinct from one another, meaning the variants had evolved in isolation from each other.
Polyclonal populations of K. pneumoniae were present within the samples, and the researchers identified four main genotypes. They hope to expand their study to investigate a larger range of samples in the near future. This will contribute to the creation of a strong database of circulating strains in Saudi Arabia.
As the research team state in their paper, published in Cureus, “these results may not give the true picture of antibiotic resistance in the whole country. A multi-centric study involving a larger number of isolates is needed [… and] investigations on the plasmids carrying the resistance genes would also shed light on horizontal transfer.”
References
Khdary, H.N. et al. Investigation on the genetic signatures of antibiotic resistance in multi-drug-resistant Klebsiella pneumoniae isolates from National Guard Hospital, Riyadh. Cureus 12 (11) e11288 (2020)
| article